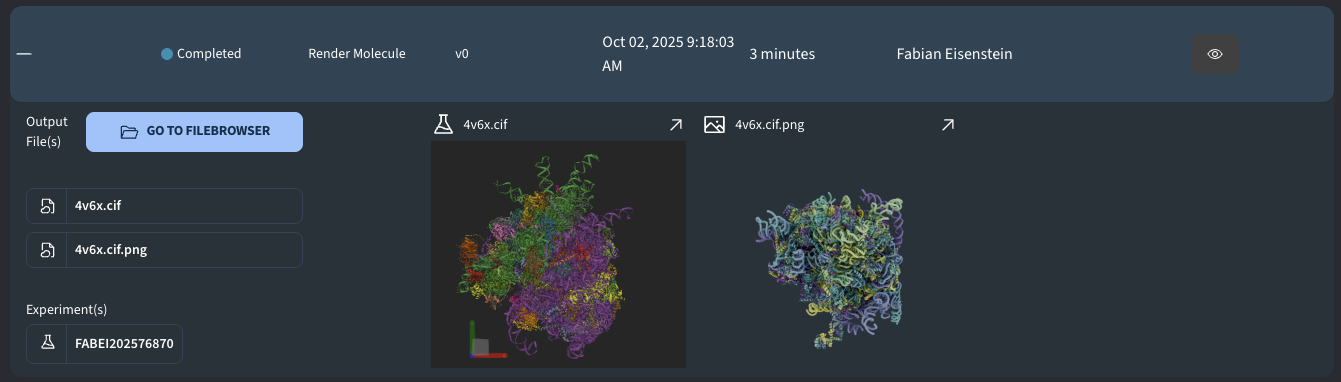
Render Molecule: Make your structure look beautiful with Blender
Use Blender and MolecularNodes to generate a beautiful render of your protein structure!
This job template is built with the Blender + MolecularNodes environment. The user must define the path to a PDB or CIF file containing a molecular structure.
The job uses a Python script that runs within blender. It loads the file, adjusts the scene and renders an image.
Quick Start
To run this job, you need to specify the following input variables:
- DC_MODEL: Path to a PDB or CIF file containing a molecular structure.
Optionally, the following input variables can also be configured:
- DC_STYLE: MolecularNodes rendering style. Can be: cartoon, ribbon, surface, spheres, sticks, ball_and_stick
- DC_COLOR_MODE: Color of the molecule. Can be: common, nc_gradient or (R, G, B, A) (e.g. "(1.0, 0.5, 0.0, 1.0)")
- DC_CMAP: CMAP gradient in case DC_COLOR_MODE is nc_gradient. Can be: any of the matplotlib cmaps
- DC_ENGINE: Renering engine. Can be: CYCLES (for ray tracing) or EEVEE (for rasterization)
- DC_RESOLUTION: Resolution in pixels of the rendered image: width and height
- DC_CENTER: Centers molecule's center of mass on center of rotation (not necessary for stills).
- DC_VIEW: Determines the side from which the molecule is viewed. Can be: default, front, back, top, bottom, left, right, all
- DC_MOVIE: Renders 360 degree turnable animation of molecule rather than still image.
- DC_MOV_SECONDS: Time in seconds to complete the 360 degree rotation.
Examples
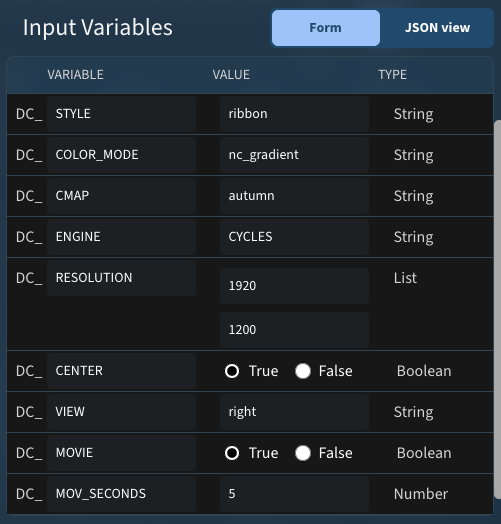
360 degree movie:
These variables run on PDB 7WAH resulted in this movie:
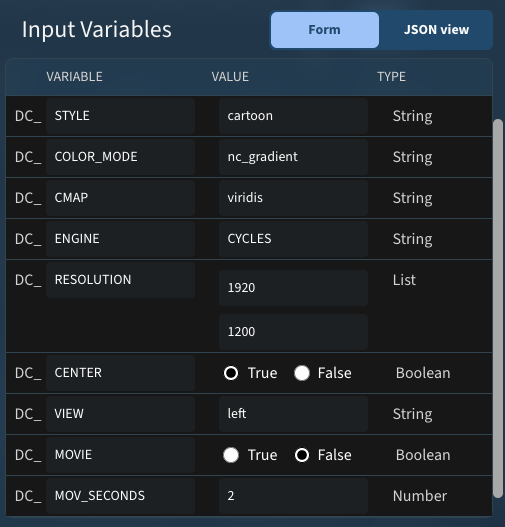
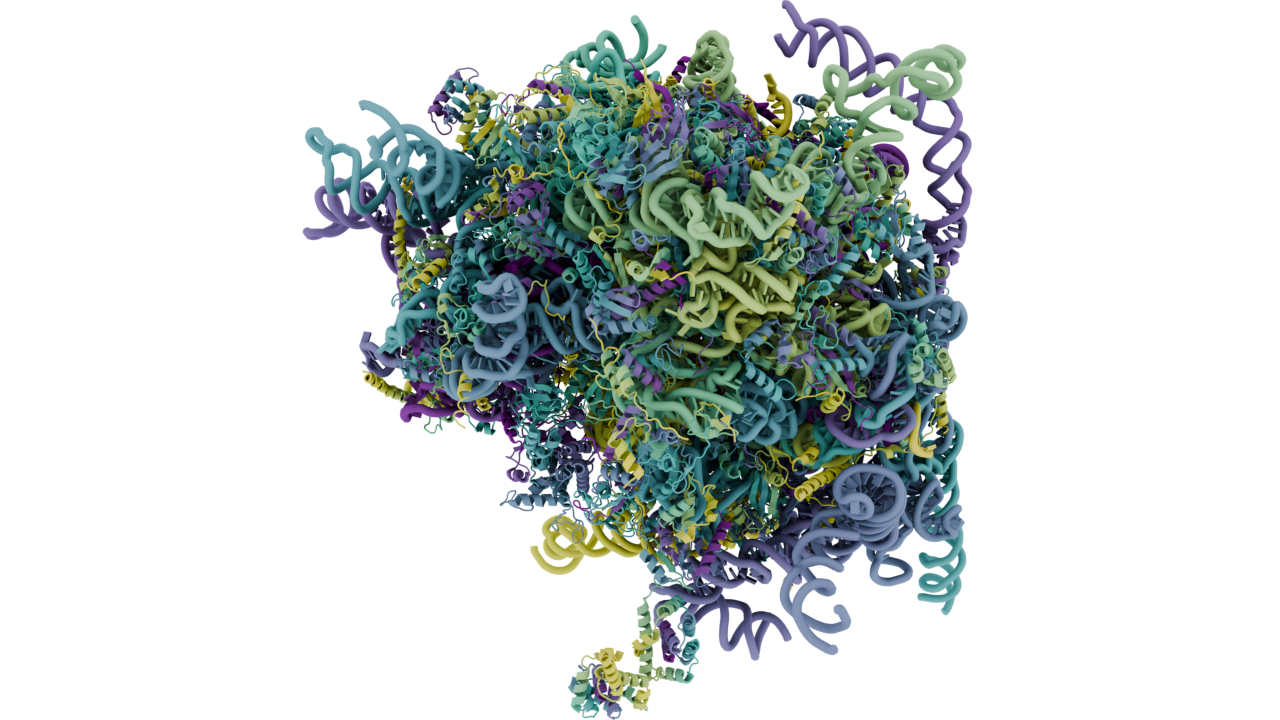
Still image:
These variables run on PDB 4V6X resulted in this image:
Output
A successfully completed job generates the following outputs: