CCP4 & XDS & PyMOL GUI
Introduction
The public CCP4 & XDS & PyMOL GUI environment supports macromolecular crystallography (MX) workflows by providing a complete set of tools for data processing, structure solution, and visualization. It includes:
- XDS for peak integration,
- generate_XDS.INP, XDSGUI, XDSVIEWER, XDSSTAT for supporting XDS workflows,
- CCP4 suite for structure solution, refinement, and visualization,
- PyMOL for additional visualization,
It is optimized for users preferring a graphical workflow while maintaining full access to command-line tools and scripting capabilities.
Environment configuration
Installation directories
- XDS binaries and tools are installed in
/opt/xds/, including:xds,xds_par,generate_XDS.INP,xdsgui,xdsviewer, andxdsstat.- Plugins for HDF5 data:
dectris-neggia.soanddurin-plugin.so.
- CCP4 is installed in
/opt/ccp4/, and its environment is automatically sourced on terminal startup. - Python virtual environment for plotting is located in
/opt/plotting/.
Python plotting venv
Python libraries for plotting results have been installed in the "plotting" venv. Installed modules include: numpy, matplotlib, h5py, hdf5plugin, argparse, pandas
Activate the venv via:
source /opt/plotting/bin/activateUsage example
XDS
The XDS setup in this environment is configured similarly to the standalone XDS environment. Please refer to the dedicated XDS help page for comprehensive usage instructions, including generating input files, configuring plugins for .h5 files, and running data processing with xds_par.
XDSGUI
XDSGUI is user-friendly interface for managing XDS jobs, viewing logs, and launching processing tasks. It supports drag-and-drop of images and integrates with standard XDS.INP workflows.
Launch with by starting a terminal and typing:
xdsgui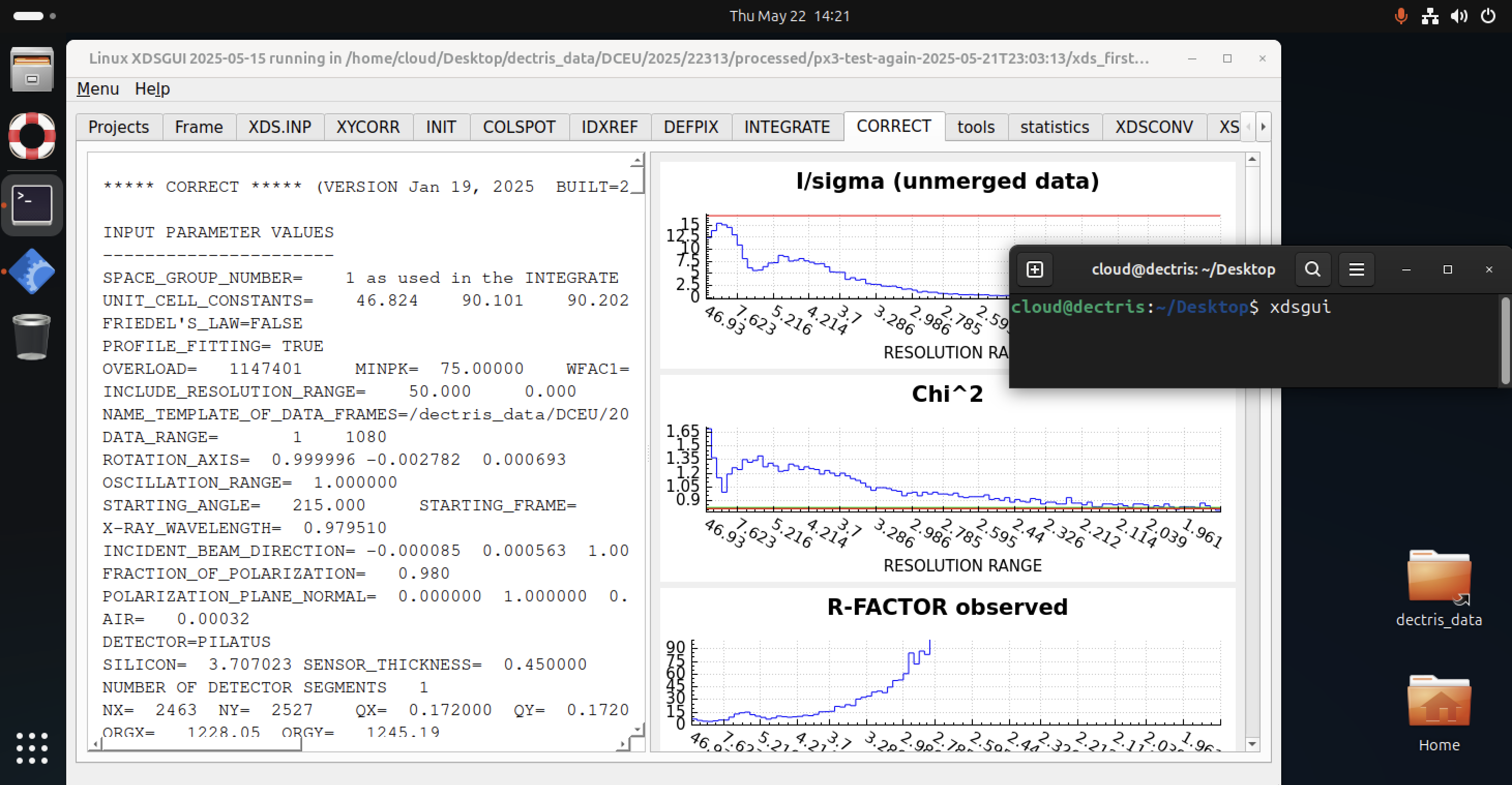
XDSSTAT
XDSSTAT can be used to evaluate the output from XDS. It generates several .pck image files containing quality statistics and can be run from the command line using:
xdsstat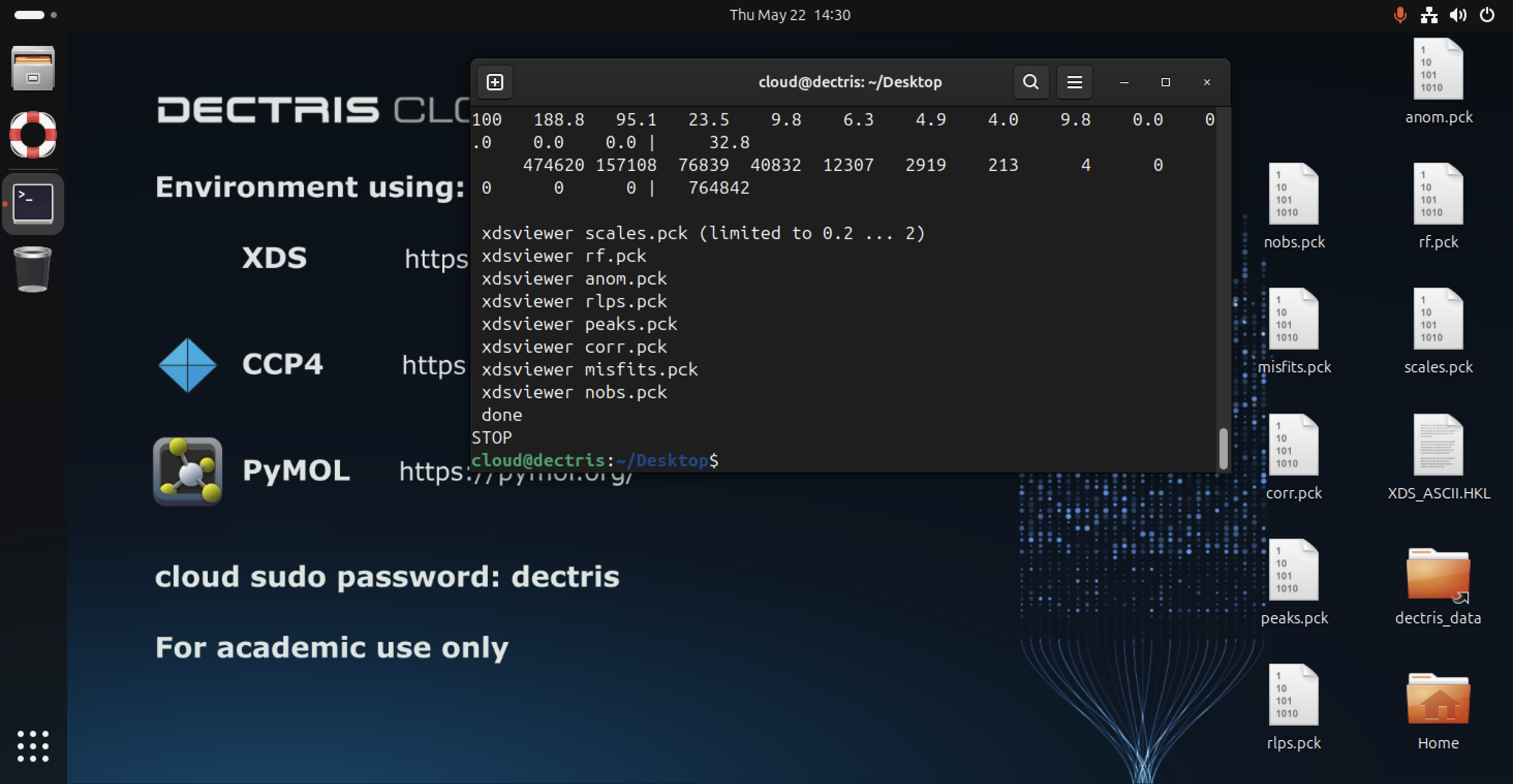
XDSVIEWER
XDSVIEWER is a graphical tool for visualizing diffraction images and processed data from XDS (including .pck image files generated with XDSSTAT). It helps users inspect spot finding, indexing, and integration results. You can launch it directly from the command line with:
xdsviewer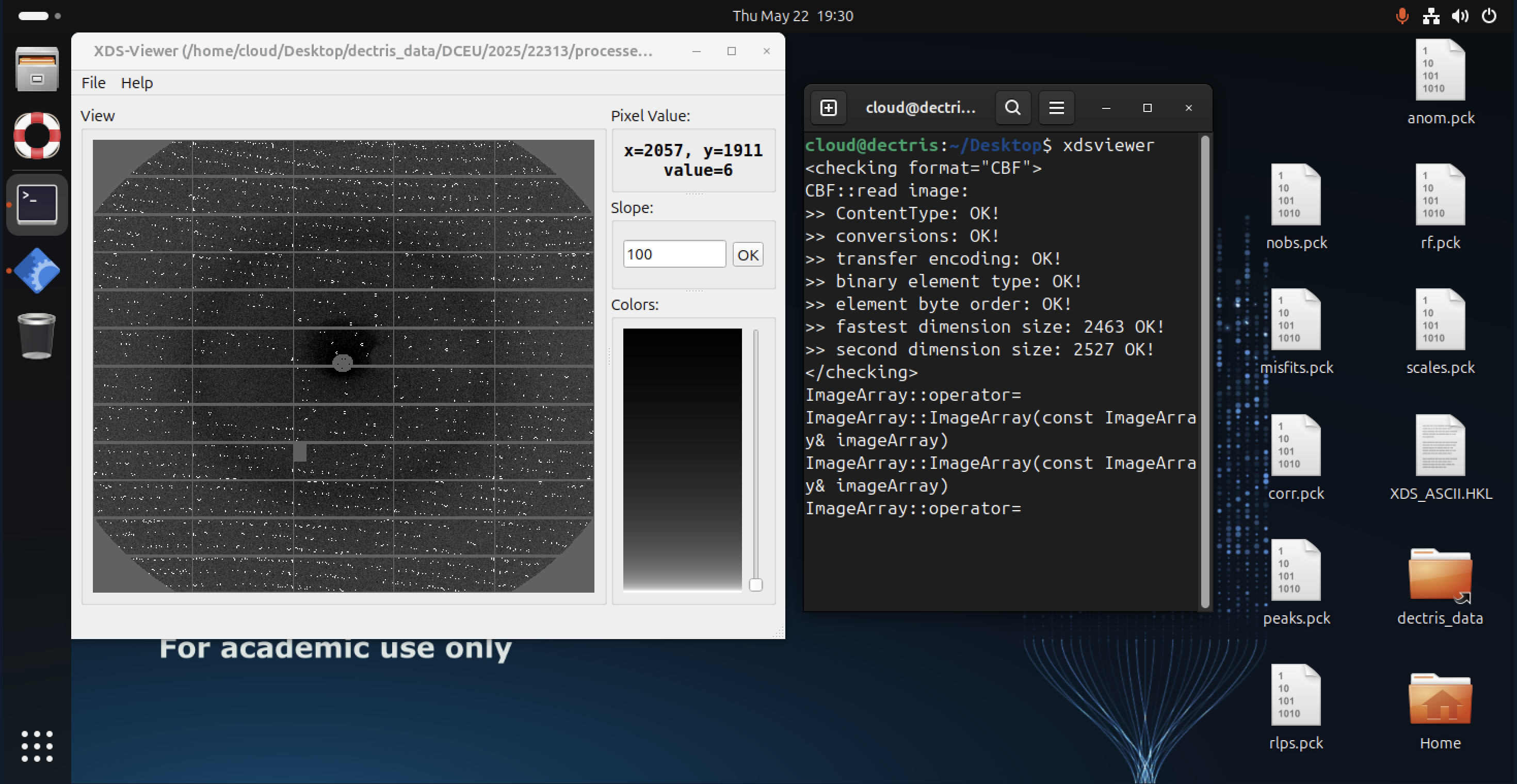
CCP4
The CCP4 suite provides a comprehensive set of tools for structure determination, refinement, and validation. You can interact with CCP4 either through its graphical interface (CCP4i2) or by using individual CCP4 programs directly from the command line.
To start the CCP4 graphical interface, open a terminal and type:
ccp4i2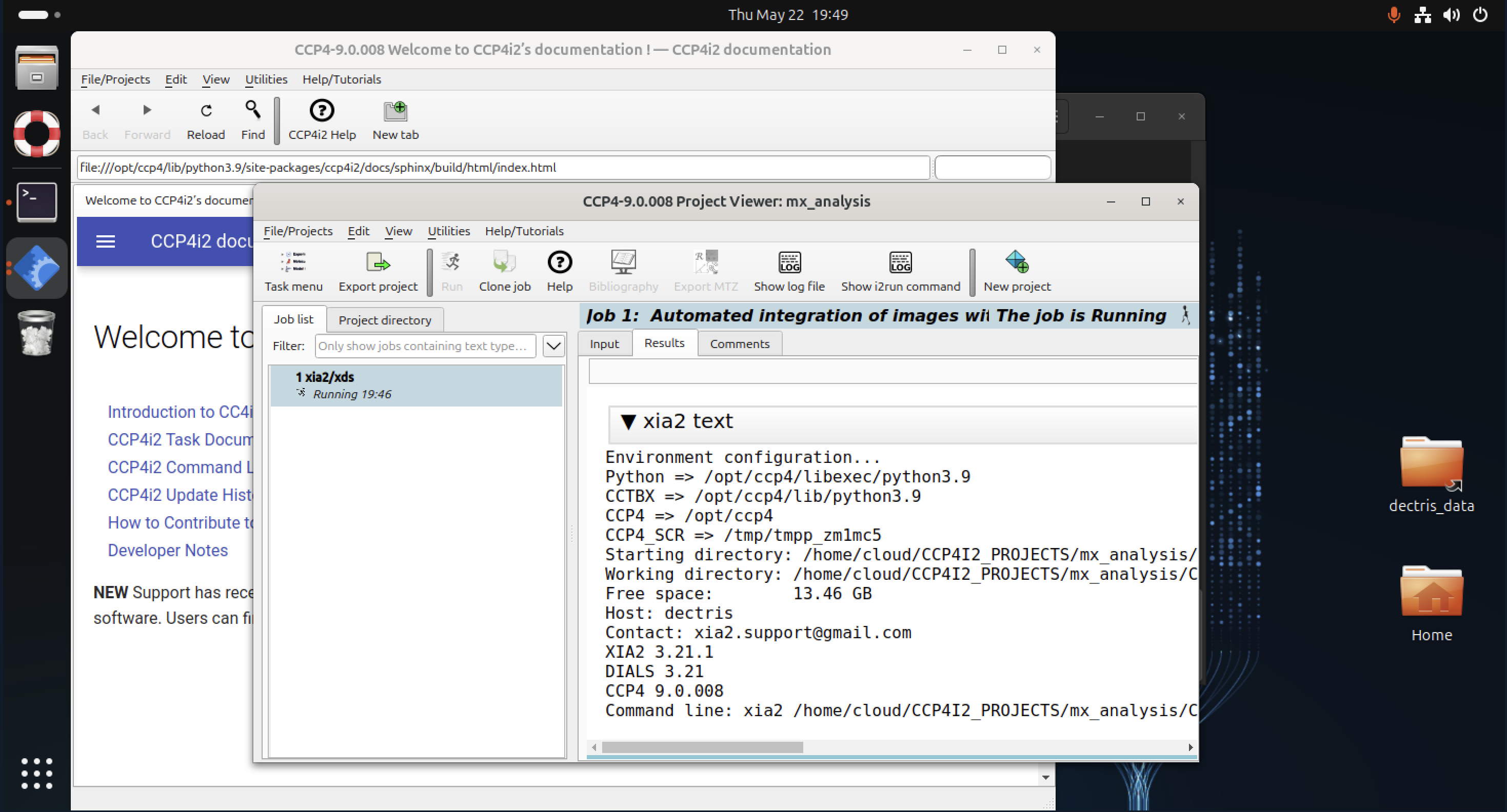
The CCP4 GUI allows easy navigation and management of projects, offering interactive workflows for tasks such as molecular replacement, experimental phasing, and refinement.
You can also use CCP4 programs individually from the terminal. After opening a terminal session, you can execute programs directly by typing their names followed by required input parameters. For example, to run pointless to analyze symmetry and indexing of reflection data:
pointless HKLIN mydata.mtz HKLout sorted.mtzSimilarly, other common programs such as dials, xia2, aimless, phaser, refmac5, or coot can be invoked from the command line:
For details on specific programs and their command-line parameters, refer to the CCP4 program documentation.
PyMOL
The PyMOL GUI can be started by opening a terminal and typing pymol:
Alternatively, PyMOL can be used in a scripting context. In the example below the python PyMOL module is imported via from pymol import cmd in a python script pymol_plot.py, which is executed from the command line via:
python3 pymol_plot.py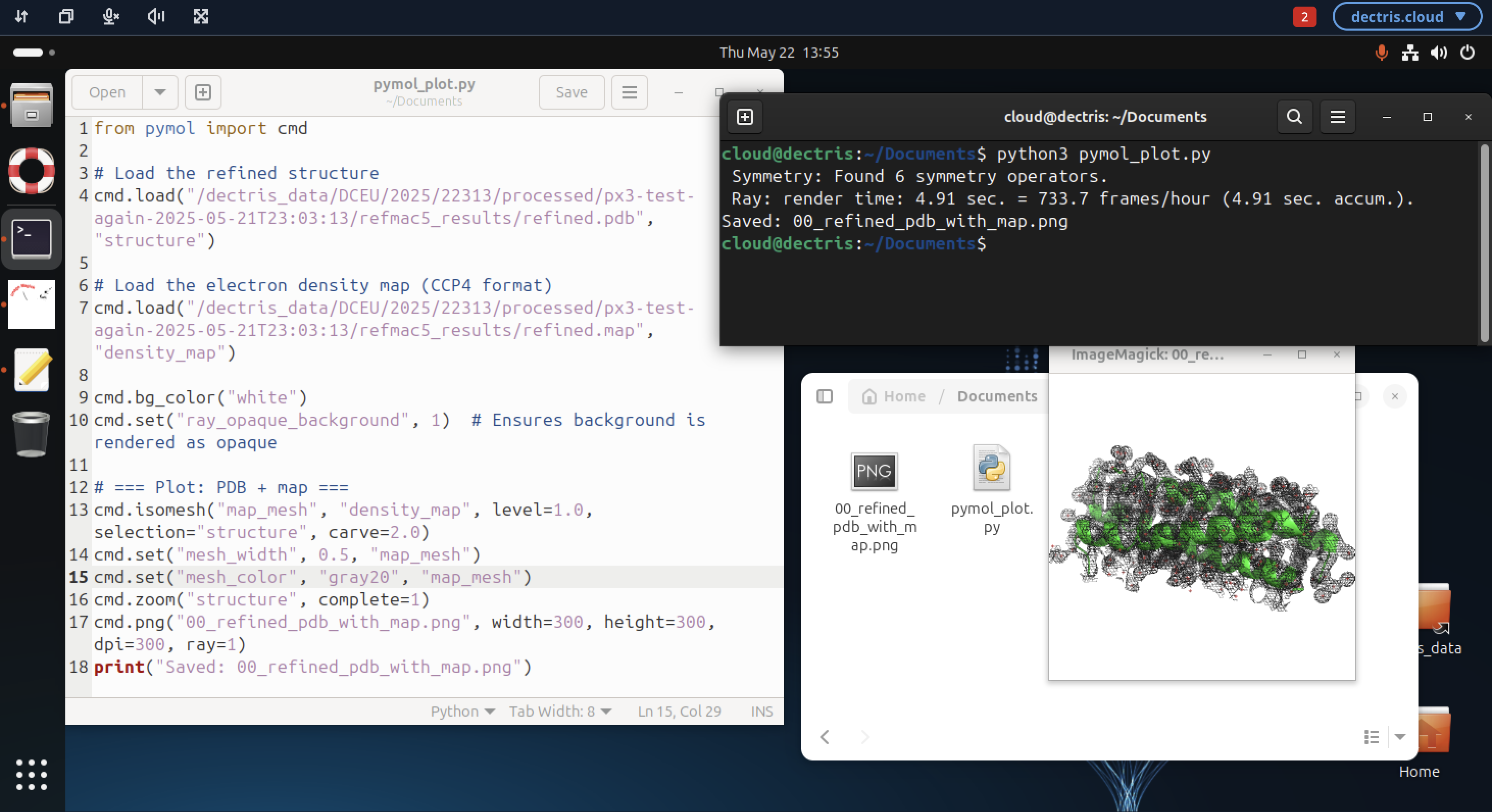
Configuring job templates
To ensure your custom scripts or templates work correctly:
- Load XDS and CCP4 tools
export PATH="/opt/xds:/opt/xds/xdsviewer:$PATH"
. /opt/ccp4/bin/ccp4.setup-sh- Optionally enable the Python plotting environment:
source /opt/plotting/bin/activatePublic job templates
The following job templates have been created for this environment and made public:
- xia2 example job: Run the xia2 pipeline from the CCP4 suite with configurable input parameters.
Version history
Version 0
Initial container with:
- XDS (version Jan 19, 2025 BUILT=20250409), and HDF5 plugins
- XDSGUI, XDSVIEWER, and XDSSTAT
- CCP4 suite (v9.0.008)
- PyMOL
- Plotting environment:
numpy,matplotlib,h5py,hdf5plugin,argparse,pandas
Version 1
- Additionally installed hdf5-tools for h5 files support.
Version 2
- Identical to version 1. Was rebuilt due to a bug to make sure the environment also works in the US region. Users with sessions in the US region should use this version and not earlier ones.
Version 3
- Updated XDS (VERSION Jan 19, 2025 BUILT=20250714)
- Updated CCP4 (v9.0.010)
References
XDS
- XDS program package: https://xds.mr.mpg.de/
- XDS references: https://xds.mr.mpg.de/html_doc/references.html
- XDS wiki: https://wiki.uni-konstanz.de/xds/index.php/Main_Page
CCP4
- CCP4 page: https://www.ccp4.ac.uk/
- CCP4 wiki: https://wiki.uni-konstanz.de/ccp4/index.php/Main_Page
- General CCP4 program reference: J. Agirre et al. Acta. Cryst. D79, 449-461 (2023) “The CCP4 suite: integrative software for macromolecular crystallography” [doi:10.1107/S2059798323003595]
- Specific CCP4 program reference info: https://www.ccp4.ac.uk/ccp4-v7-0-program-references/
PyMOL
- PyMOL page: https://pymol.org/
- PyMOL wiki: https://pymolwiki.org/index.php/Main_Page
- PyMOL citation info (bottom of page): https://www.pymol.org/support.html?
