xia2 example job: Automated MX indexing, integration & scaling
Introduction
xia2 is an automation layer for macromolecular (and small-molecule) crystallography data reduction. Given a folder of raw diffraction images, it runs the full processing pipeline from indexing and integration to scaling and merging, orchestrating engines such as DIALS and XDS and handling symmetry determination with POINTLESS/AIMLESS in CCP4. The result is a consistent, reproducible pipeline that produces merged data (MTZ), logs, and a rich HTML report.
This example job template exemplifies how xia2 can be run as a job on DECTRIS CLOUD with configurable input and machine performance types. The job template has been built with the CCP4 & XDS & PyMOL command line environment. You point it to an image directory, choose a pipeline (dials, dials-aimless, 3d/xds, or 3dii), and the template:
- builds the appropriate
xia2command from your inputs (space group, unit cell, resolution limits, optional anomalousatom=…, and DIALS small-molecule mode), - runs the processing and collects the standard xia2 output,
- generates summary plots (CC½, I/σ(I), completeness, multiplicity, etc.),
- and records simple CPU/Memory usage over time.
For XDS pipelines, an HDF5 plugin (dectris-neggia.so) is provided as part of the environment and is included in the xia2 call.
Note that this job template is based on an environment that is only available for academic users.
Quick Start
To run this job, valid input for the following variables are required to be defined:
- DC_PATH: Path to a folder within your experiment or project that contains your diffraction images. If you give relative image names, they’re resolved against this path.
Example:/dectris_data/DECTRIS/2025/23393/raw/ALS831_pilatus_1/data/
Optionally, the following input variables can also be configured. If left with a value of -1, the input will be ignored within the job:
- DC_IMAGE: Makes it possible to specify the image input option for dials, for example to specify a specific dataset within a folder or a specific range of images to use. Up to five strings can be defined:
Example:data_J1_0001.cbf:1:200 - DC_PIPELINE: Which backend to use.
Allowed values:dials(DIALS + dials.scale),dials-aimless(DIALS + AIMLESS),3d(XDS),3dii(XDS all-frames peak search) - DC_SPACEGROUP: Specify a space group (otherwise let xia2/pointless decide).
Example:P212121 - DC_UNIT_CELL: Specify a unit cell as six comma-separated values
a,b,c,alpha,beta,gamma.
Example:78.3,94.1,101.2,90,90,90 - DC_D_MIN: High-resolution cutoff in Å.
Example:2.2 - DC_D_MAX: Low-resolution cutoff in Å.
Example:50 - DC_MON_INTERVAL: Resource monitor sampling interval in seconds.
Default:30
Example:10 - DC_ATOM: Option to separate anomalous pairs.
Example:X - DC_SMALL_MOLECULE: Enable small-molecule mode (DIALS pipelines only:
dials,dials-aimless). Ignored for XDS pipelines (3d,3dii).
Example:true
Output
A successfully completed job generates the following outputs:
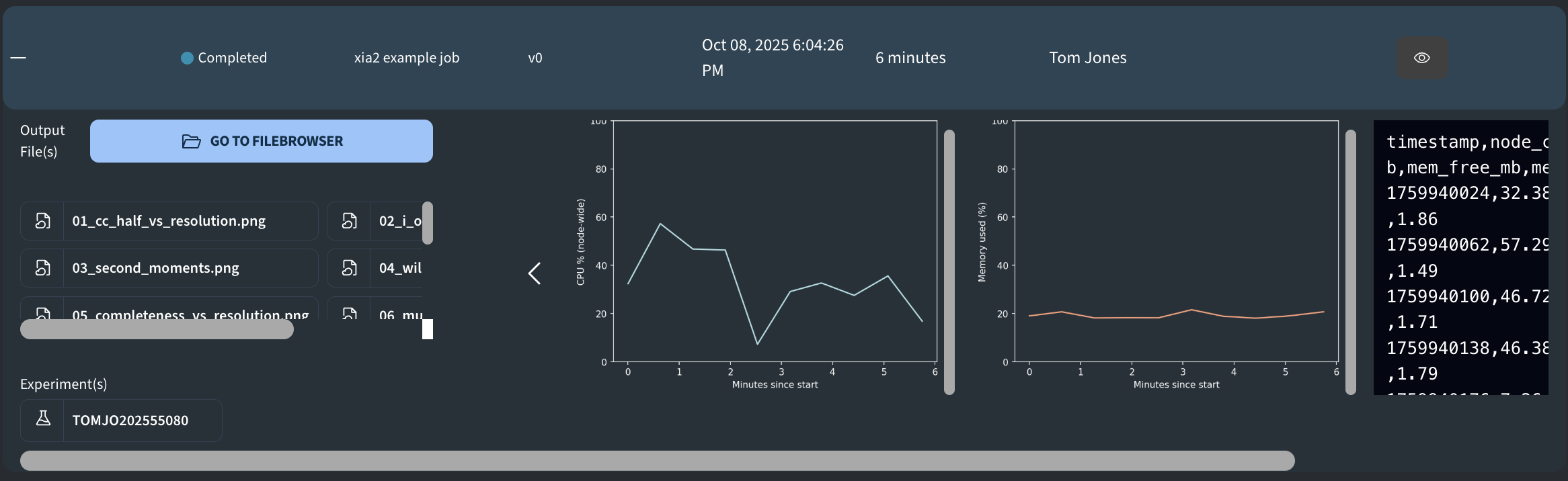
At the top level of the output folder, the following plots and files are defined:
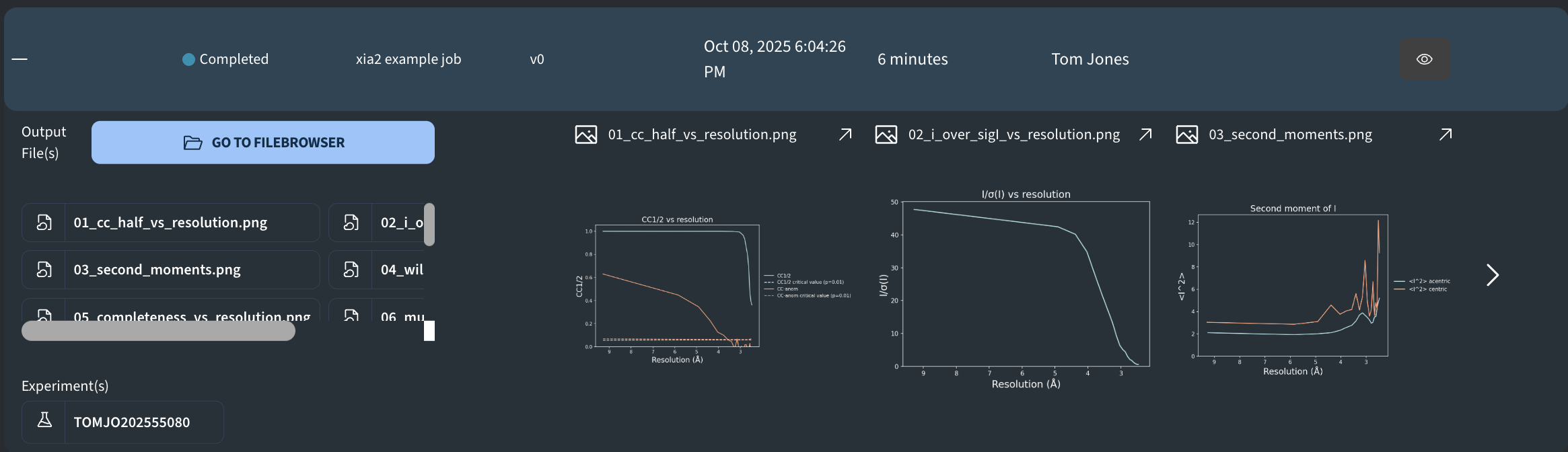
- 01_cc_half_vs_resolution.png: CC½ and CC-anom vs resolution (Å), with p=0.01 critical curves overlaid..
- 02_i_over_sigI_vs_resolution.png: Mean I/σ(I) vs resolution (Å): average signal-to-noise per bin.
- 03_second_moments.png: Second moment vs resolution (Å).
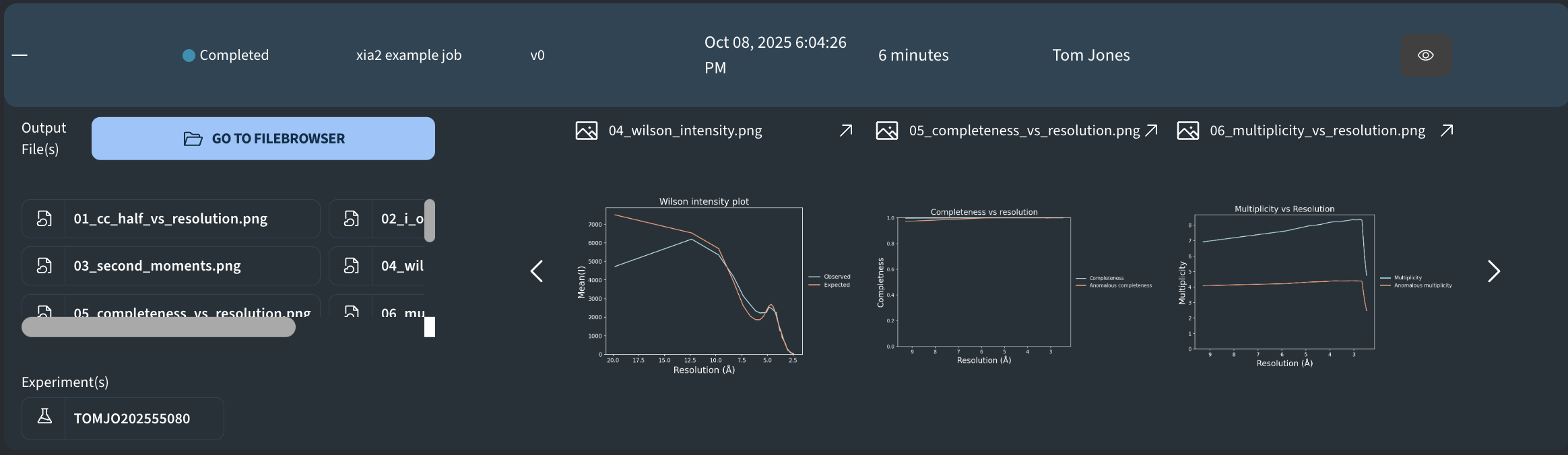
- 04_wilson_intensity.png: Wilson intensity plot (mean intensity vs resolution).
- 05_completeness_vs_resolution.png: Completeness (%) vs resolution (Å).
- 06_multiplicity_vs_resolution.png: Multiplicity (redundancy; observations per unique) vs resolution (Å).
- cpu_total_vs_time.png: Node-wide CPU utilization (%) over job runtime.
- mem_used_pct_vs_time.png: Node-wide memory used (%) over job runtime
- resource_usage.csv: File used for tracking the resource usage during the job runtime. The file contains the following columns:
timestamp(epoch time),node_cpu_pct,mem_used_mb,mem_free_mb,mem_used_pct,load1(1-minute load average). - resource_usage_with_mins.csv: Same as above plus an extra column
mins_since_start, calculating the run time in minutes. - xia2-citations.txt: Relevant citations when generating results using xia2.
- xia2-summary.txt: Overall results summary.
- xia2.html: Full interactive xia2 report.
- xia2_command.txt: The final xia2 command constructed by the job, based on your given input parameters.
Within the subdirectory xia2_output, the typical xia2 output files can be found, including the subdirectories DEFAULT, DataFiles, and LogFiles.
Example input configurations
Giving only the data source as input
A minimal configuration of the job template involves configuring just DC_PATH and leaving all other parameters at their default value:
resulting in the following xia2 command to be constructed and called within the job template:
xia2 pipeline=3d /dectris_data/TOMJO/2025/55080/raw/data_1/ hdf5_plugin=/opt/xds/neggia.soUsing a subset of images
Sometimes you may want to process only part of a dataset, for example the early frames to minimize radiation damage. In the example below, only the first 500 images are included in the analysis, by specifying xrd_data_0001.img:1:5000 for the first entry of DC_IMAGE:
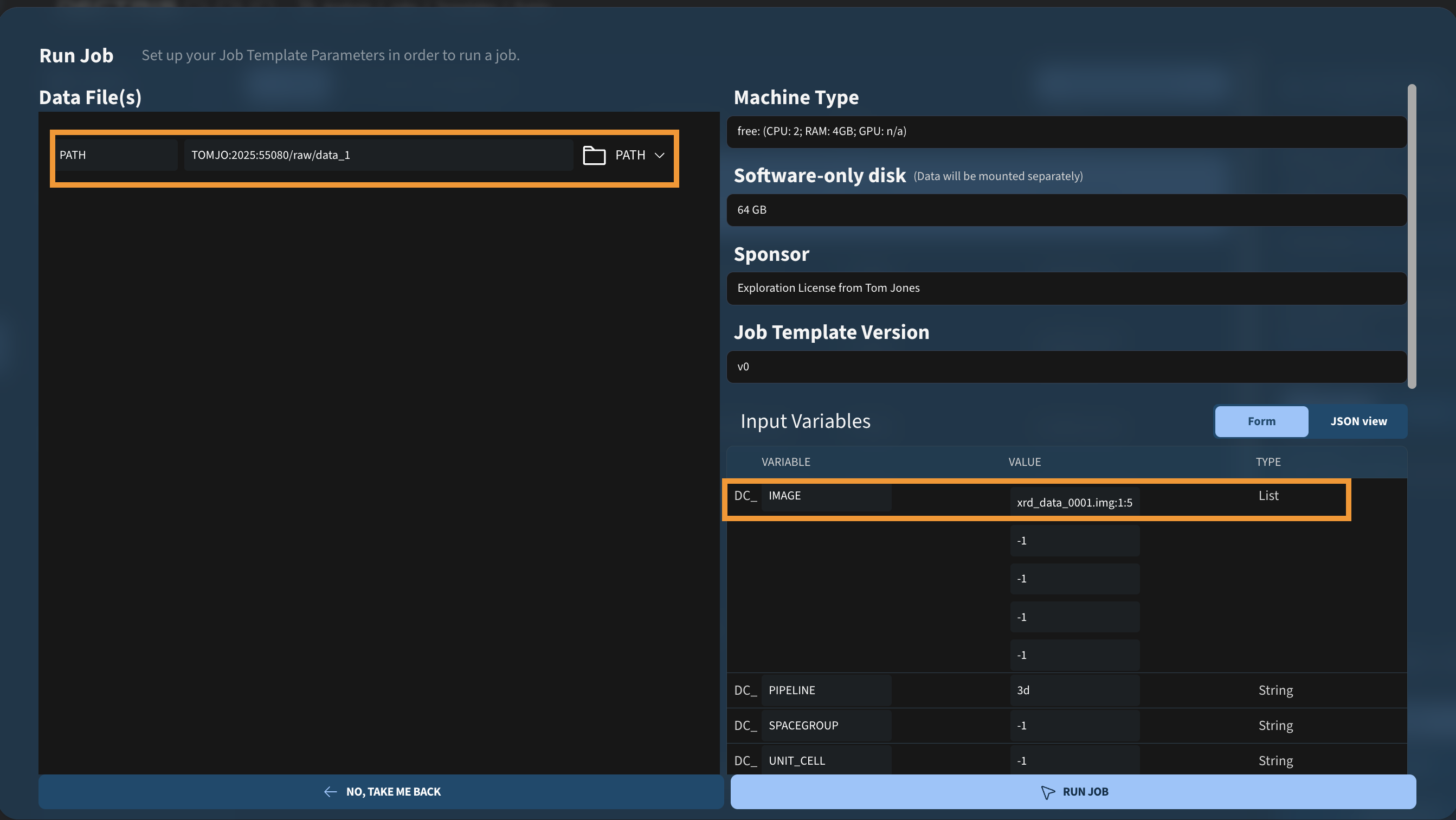
The configured string is appended to DC_PATH, resulting in the following xia2 command:
xia2 pipeline=3d image=/dectris_data/TOMJO/2025/55080/raw/data_1/xrd_data_0001.img:1:500 hdf5_plugin=/opt/xds/neggia.soSpecifying the cell parameters and unit cell
If you already know the crystal geometry (from a reference dataset or prior refinement), you can fix it for the run:
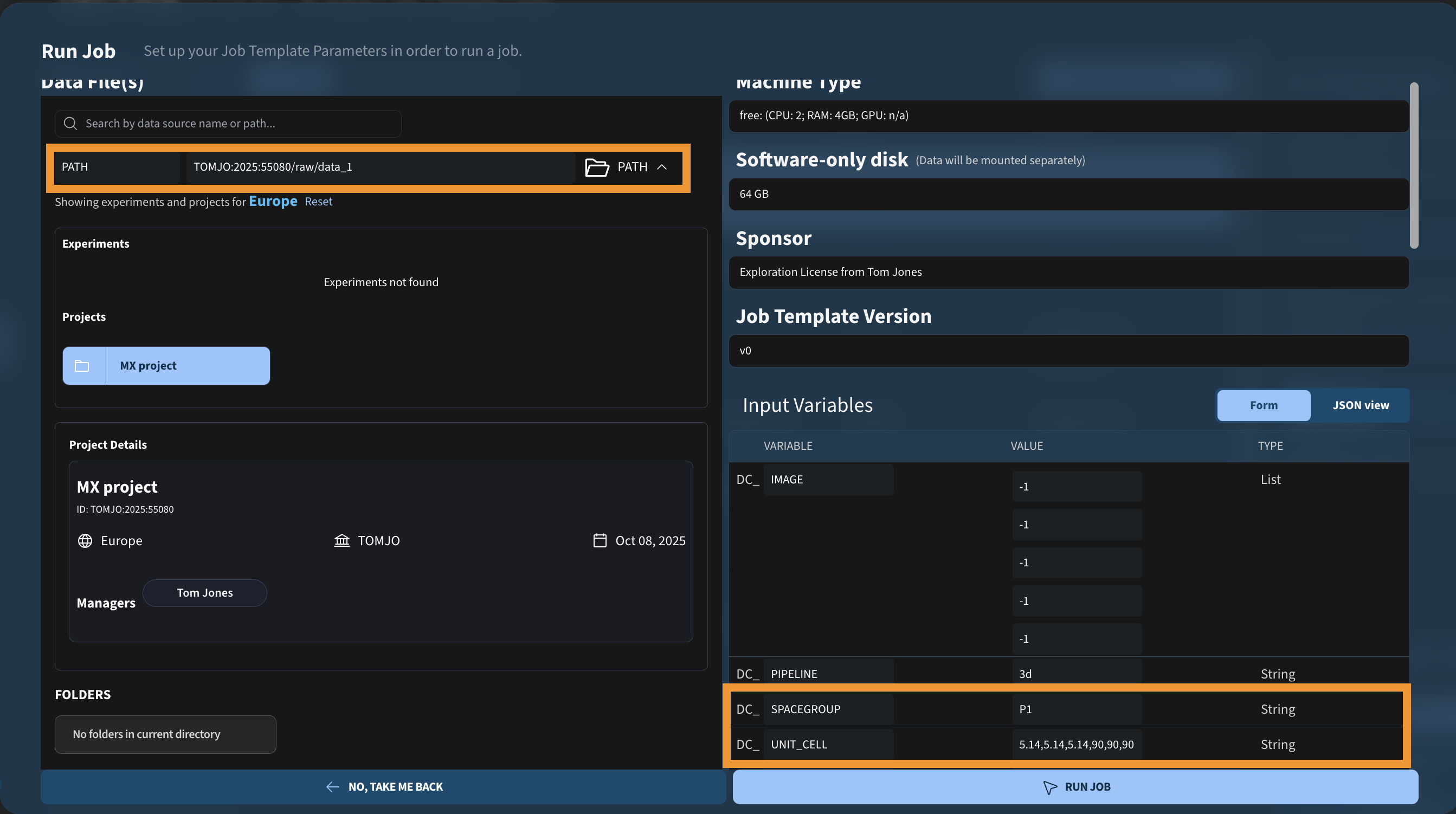
resulting in the following xia2 command:
xia2 pipeline=3d /dectris_data/TOMJO/2025/55080/raw/data_1/ xia2.settings.space_group=P1 unit_cell=5.14,5.14,5.14,90,90,90 hdf5_plugin=/opt/xds/neggia.soVersions
Version 0
- Initial version of the script, built with the version 2 of the CCP4 & XDS & PyMOL command line environment.