Molecular Replacement MOLREP
Introduction
MOLREP is a program from the CCP4 suite for performing molecular replacement in macromolecular crystallography. Given an experimental dataset and a suitable reference model, it determines the orientation and position of the model in the unit cell.
This job template illustrates how MOLREP can be run as a job on DECTRIS CLOUD using a configurable command-line workflow. The template is built on the CCP4 & XDS & PyMOL command line environment and is executed in a non-interactive batch setting.
You provide an input MTZ file containing structure-factor amplitudes and a PDB code identifying a reference model. The template then:
- prepares the CCP4 runtime environment,
- downloads the reference structure from the Protein Data Bank (PDB),
- runs MOLREP with configurable structure-factor labels and optional parameters such as the number of monomers, solvent fraction, and sequence information,
- collects the resulting models, logs, and intermediate files into the job output directory.
The result is a reproducible molecular replacement workflow that can be reused across datasets by adjusting only the job input parameters.
Quick Start
To run this job, valid input for the following variables is required:
- DC_MTZ_IN: Path to an MTZ file containing structure-factor amplitudes and uncertainties.
Example:/dectris_data/<project>/<experiment>/processed/processed_dir/merged.mtz - DC_PDB_CODE: Four-character PDB identifier of the reference model to be used for molecular replacement.
Example:1ABC - DC_F_LABEL: Column label for structure-factor amplitudes (F) in the MTZ file.
Default:F - DC_SIG_LABEL: Column label for uncertainties (SIGF) in the MTZ file.
Default:SIGF - DC_N_MONOMERS: Expected number of monomers in the asymmetric unit.
Default:0 - DC_SOLVENT_FRACTION: Solvent fraction of the crystal (e.g.
0.45).
Default:0 - DC_SEQ_FILE: Path to a sequence file (FASTA format).
Default:/path/to/seq.fasta$
Output
A successfully completed job produces the following output files in the job output directory:
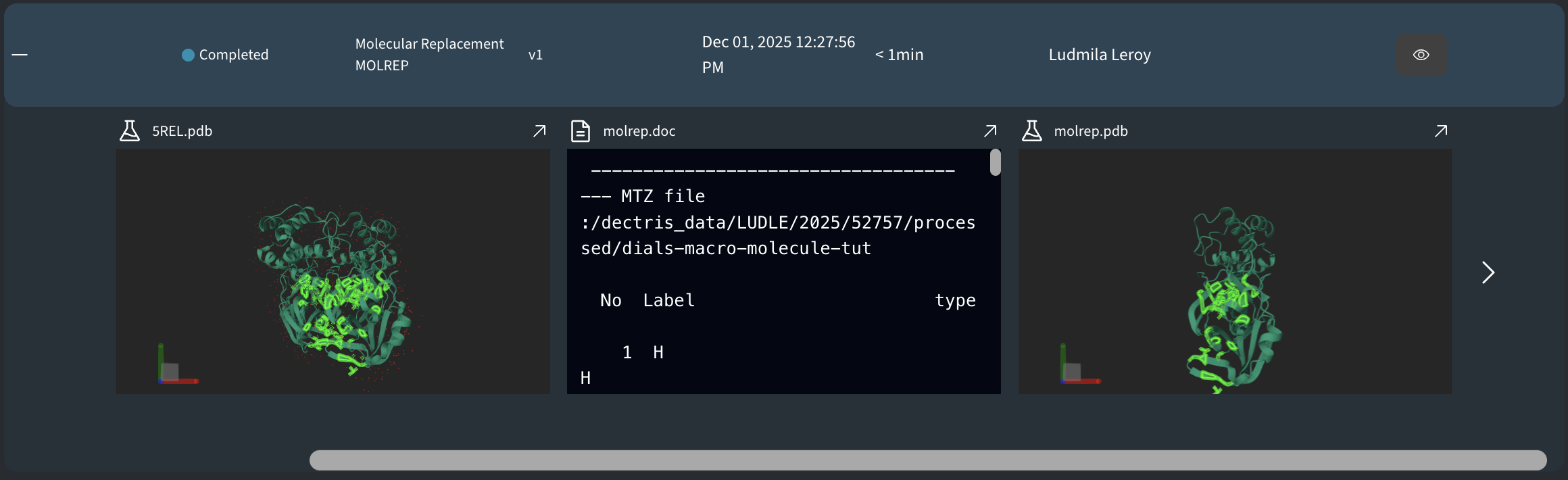
- 1ABC.pdb: The reference model downloaded from the Protein Data Bank (PDB) using
DC_PDB_CODE. This is the input search model provided to MOLREP. - molrep.pdb: The molecular replacement solution produced by MOLREP (placed and oriented model in the target unit cell).
- molrep.doc: MOLREP log/documentation file containing run parameters, MTZ label interpretation, search progress (rotation/translation results), scores, and summary statistics.
Versions
Version 1
- Adds
molrep.docto the job table output.
Built with version 3 of the CCP4 & XDS & PyMOL command line environment.
Version 0
- Initial version of the MOLREP job template.
Built with version 3 of the CCP4 & XDS & PyMOL command line environment.