Fast DP: Fast Data Processing with XDS
Introduction
The Fast DP environment is a lightweight, command-line–focused workspace for macromolecular crystallography (MX) data processing and rapid downstream analysis. It is designed for users who want to run fast_dp and related tooling in a reproducible container setup, while still having access to commonly used MX utilities for inspection, plotting, and scripting.
The environment is built on Ubuntu 24.04 and combines:
- Fast DP (via conda) for fast automated processing and summary output,
- CCTBX components (via
cctbx-base) for crystallographic functionality used by fast_dp and related tools, - XDS tools for integration workflows and supporting utilities,
- CCP4 command-line tools (GUI components removed) for standard MX utilities,
- PyMOL for structure visualization,
- common Python libraries for scripting and plotting (numpy, matplotlib, pandas, h5py, hdf5plugin).
This environment is primarily intended for scripted and automated workflows, in particular DECTRIS CLOUD job templates.
Environment configuration
Installation directories
Key software components are installed in fixed locations inside the container:
- Conda (Miniconda):
/opt/conda/ - fast_dp conda environment:
/opt/conda/envs/fastdp/
This conda environment contains:fast_dp- CCTBX components (via
cctbx-base) - Python (3.11)
- Scientific Python stack (numpy, matplotlib, pandas, h5py, hdf5plugin)
- CCTBX (via conda):
Installed inside thefastdpconda environment at:/opt/conda/envs/fastdp/
CCTBX libraries, binaries, and Python modules are available automatically when thefastdpenvironment is onPATH. No separate CCTBX installation directory is used. - XDS:
/opt/xds/
Includes:xds,xds_par- helper scripts such as
generate_XDS.INP - HDF5 plugins:
/opt/xds/dectris-neggia.so/opt/xds/durin-plugin.so
- CCP4 (command-line):
/opt/ccp4/
CCP4 is installed without GUI components and is intended for command-line use and scripting.
PATH and CCP4 setup
Interactive shells are configured to automatically:
- prepend the following directories to
PATH:/opt/conda/envs/fastdp/bin/opt/conda/bin/opt/xds
- source the CCP4 environment:
. /opt/ccp4/bin/ccp4.setup-sh
- This configuration is applied via:
/etc/profile.d/fastdp-path.sh/etc/bash.bashrc
As a result, tools such as fast_dp, CCTBX utilities, XDS programs, and CCP4 binaries are available immediately in interactive terminals.
Usage Examples
Running fast_dp command
Due to the configuration of the PATH variable, the fast_dp command can be immediately run with a path to a dataset inside a terminal:
fast_dp /path/to/data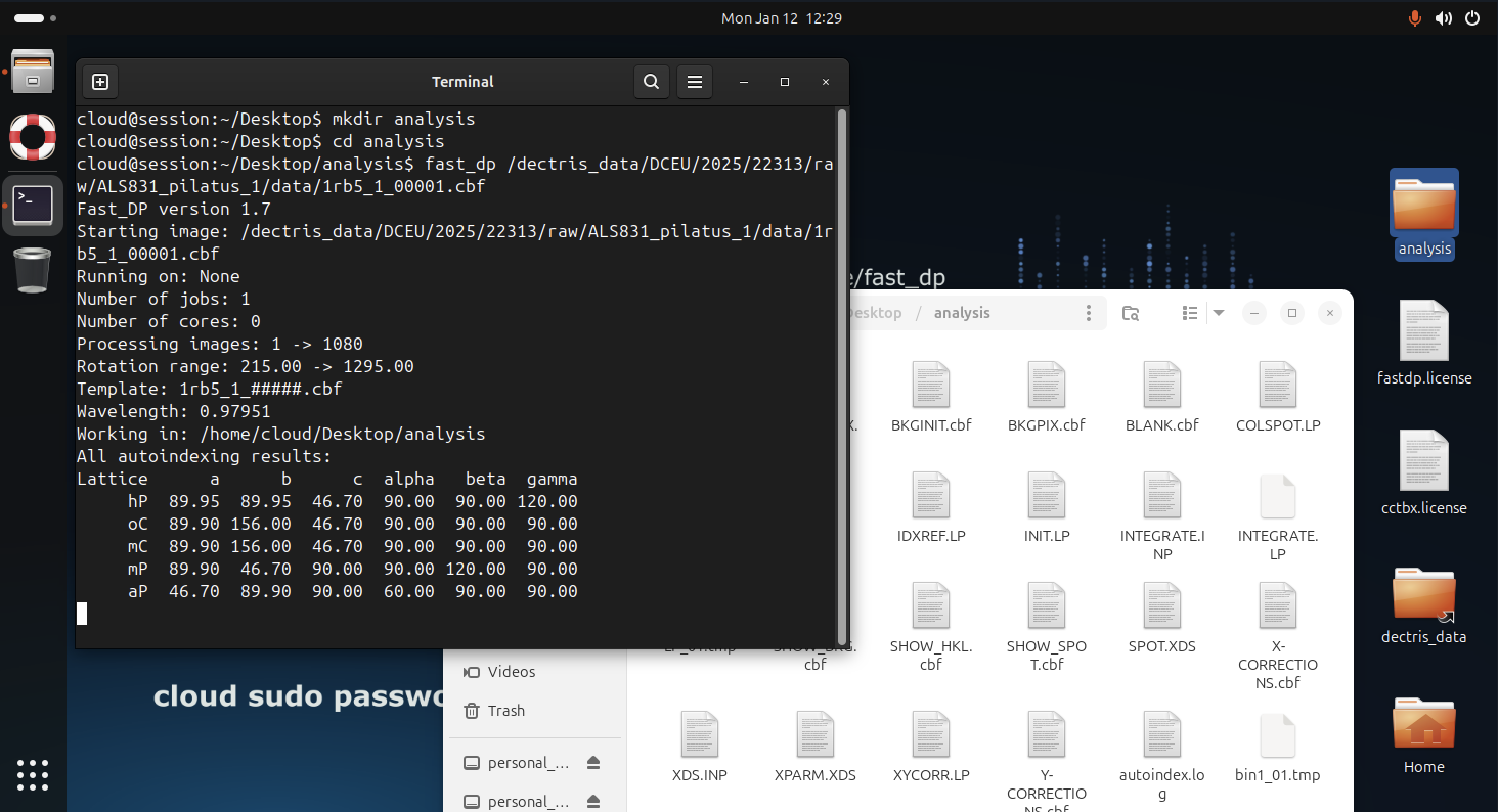
For HDF5 / NeXus input (for example EIGER detector data), one can specify a path to the DECTRIS HDF5 reader plugin:
fast_dp -l /opt/xds/dectris-neggia.so /path/to/data.h5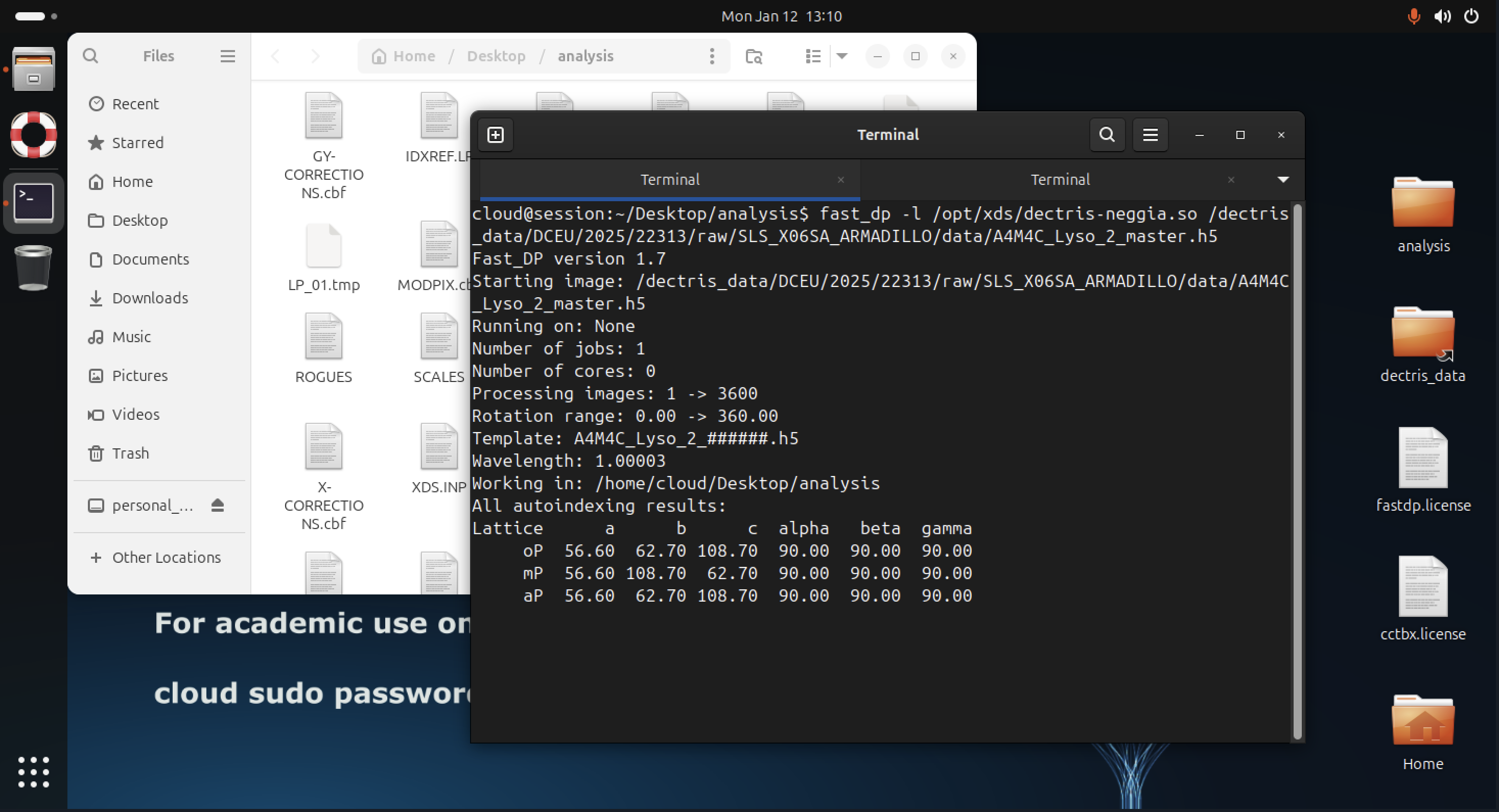
Additional fast_dp options may also be specified explicitly, for example to constrain space group or resolution:
fast_dp \
-s P212121 \
-r 2.0 \
-R 50.0 \
/path/to/data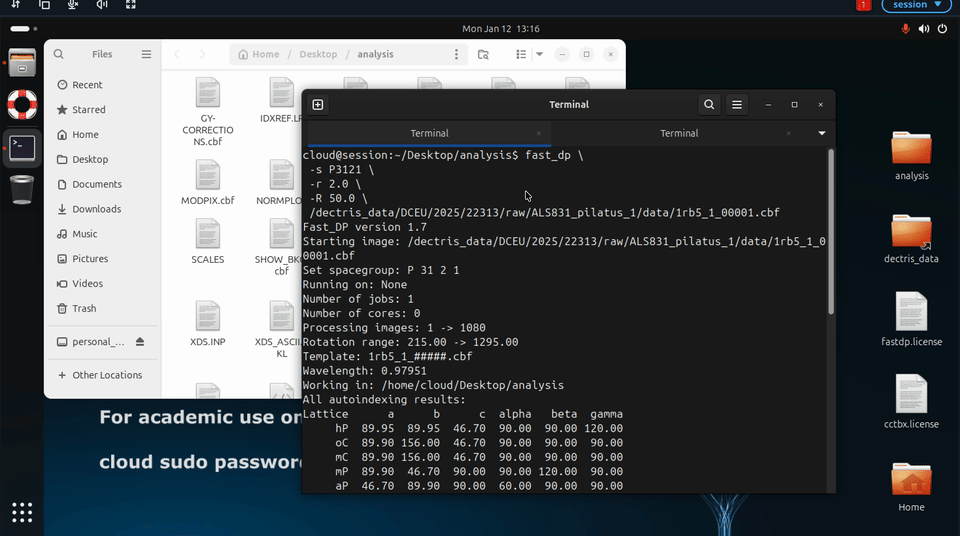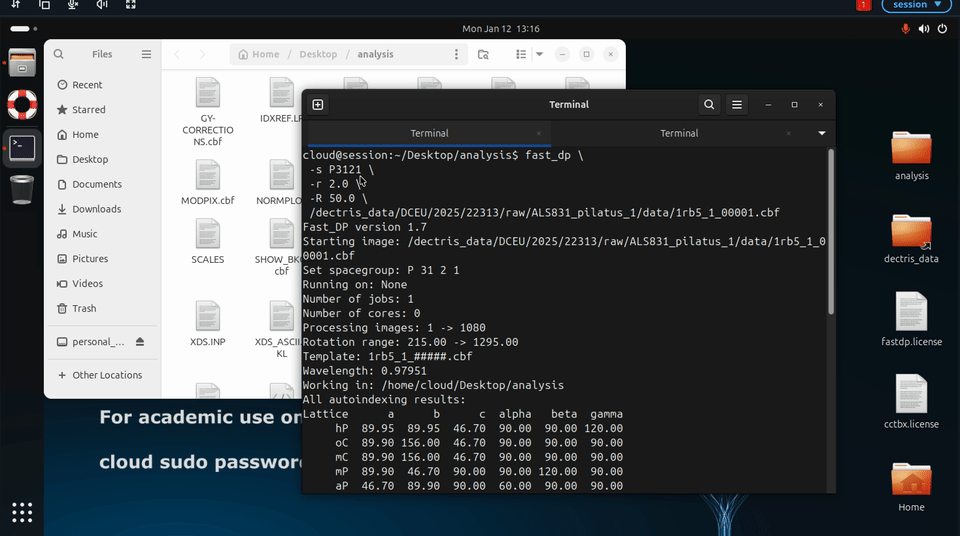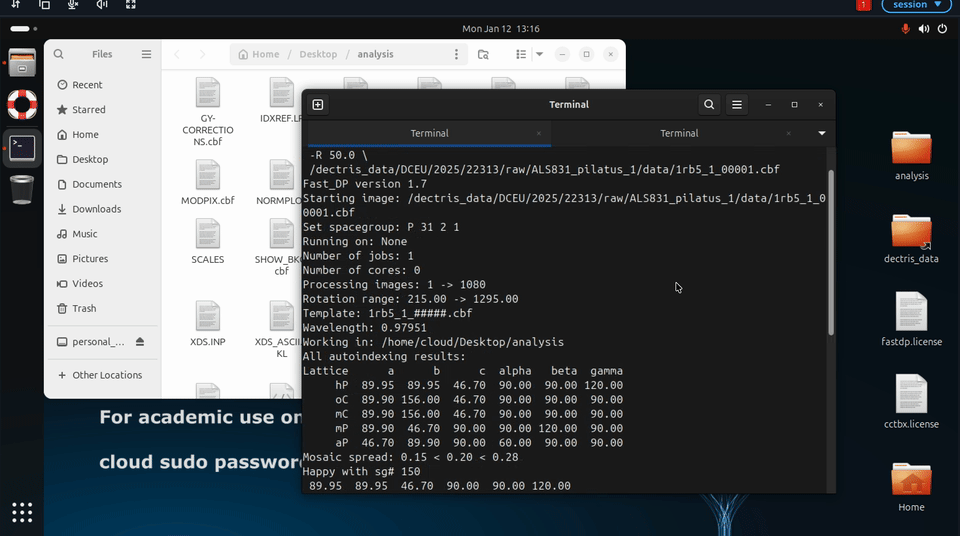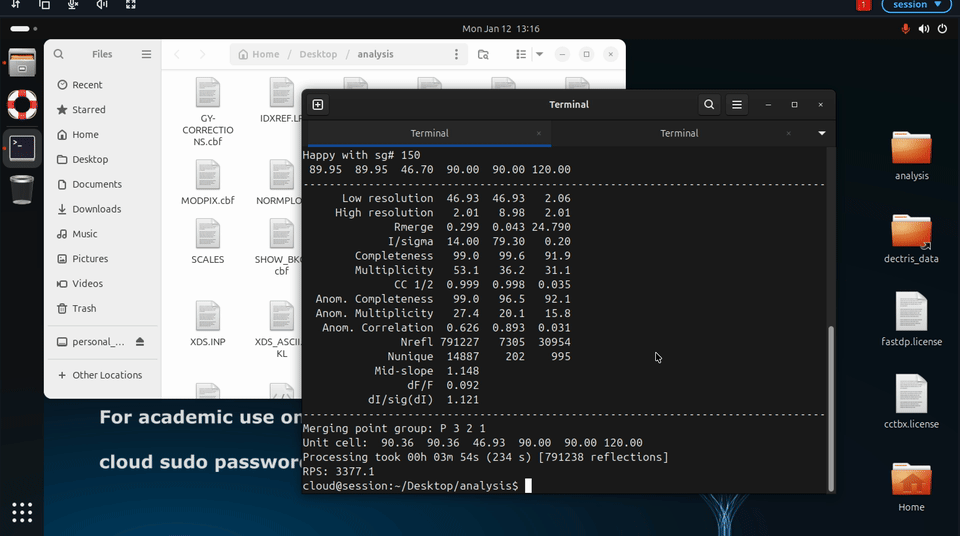
Using CCTBX for inspection and scripting
You can access CCTBX functionality from Python, for example to inspect reflection data in an MTZ file produced by fast_dp:
python3
from iotbx import reflection_file_reader
mtz = reflection_file_reader.any_reflection_file("fast_dp.mtz")
arrays = mtz.as_miller_arrays()
for arr in arrays:
print(arr.info().label_string())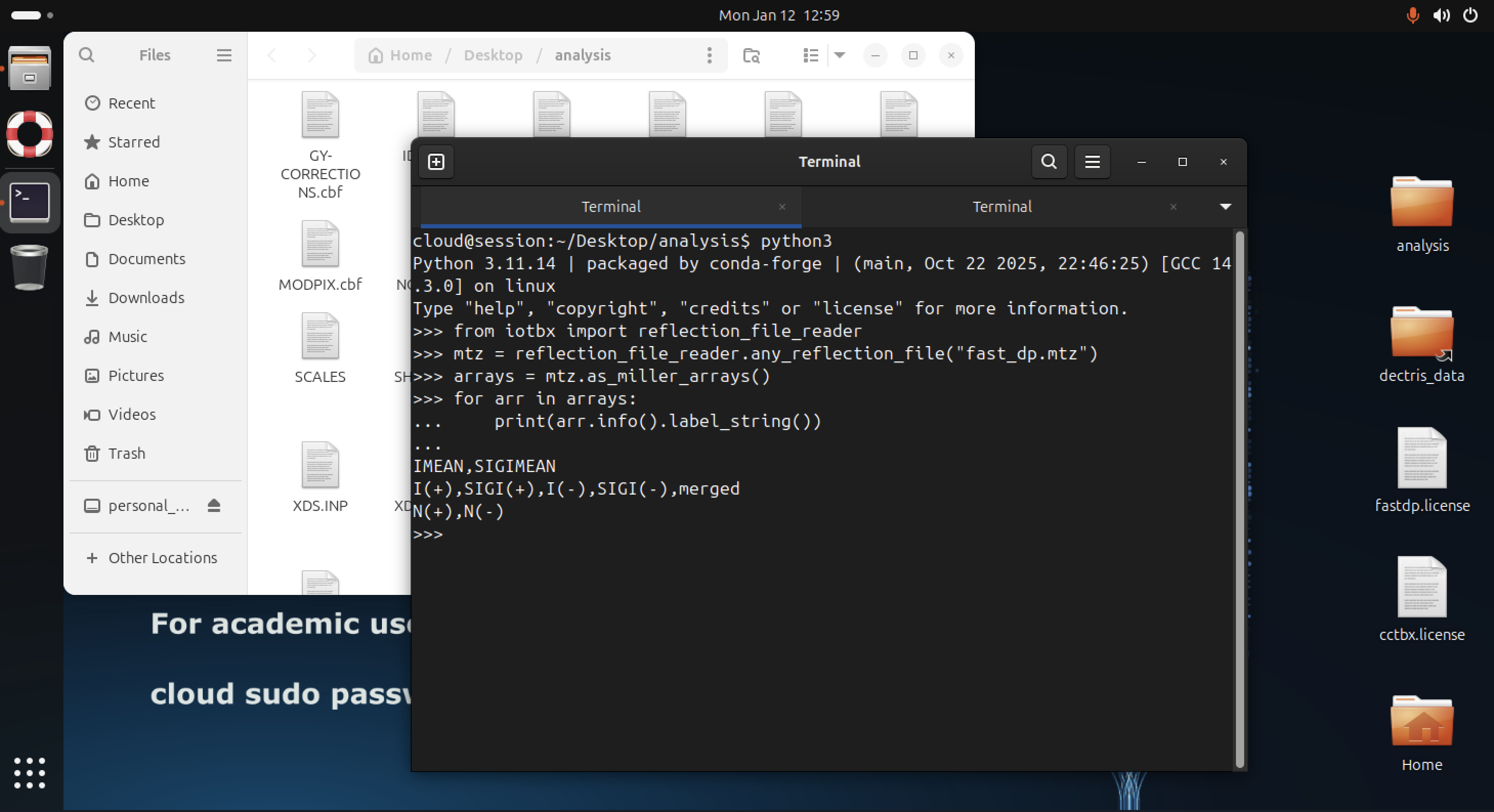
In the example above, the column labels contained in the MTZ file are printed. This pattern can be extended in Python scripts or job templates to compute statistics, inspect resolution limits, or prepare input files for downstream processing.
Configuring job templates
Job templates run in non-interactive shells and should therefore configure their environment explicitly.
At the start of your script:
# Load CCP4 command-line environment
source /opt/ccp4/bin/ccp4.setup-sh
# Make fast_dp, CCTBX, and XDS tools available export PATH="/opt/conda/envs/fastdp/bin:/opt/conda/bin:/opt/xds:$PATH"
This ensures that:
fast_dpand the associated CCTBX Python modules are available,- XDS command-line tools (e.g.
xds,xds_par) are onPATH, - CCP4 programs (e.g.
ctruncate,pointless) can be invoked directly.
Public Job Templates
Version History
Version 1
- Changes compared to Version 0:
- Added
libquadmath0to the runtime image to ensure compatibility with XDS-based workflows. - No changes were made to the installed software versions.
- Added
Version 0
- Initial release of the Fast DP command-line environment, including:
- XDS: VERSION Jan 19, 2025 (BUILT=20250714)
- CCP4: software suite version 9.0.011 (patch level 9.0.011)
- CCTBX: version 2024.2 (via
cctbx-baseconda package) - PyMOL: version 3.0.0
References
XDS
- XDS program package: https://xds.mr.mpg.de/
- XDS references: https://xds.mr.mpg.de/html_doc/references.html
- XDS wiki: https://wiki.uni-konstanz.de/xds/index.php/Main_Page
CCP4
- CCP4 page: https://www.ccp4.ac.uk/
- CCP4 wiki: https://wiki.uni-konstanz.de/ccp4/index.php/Main_Page
- General CCP4 program reference: J. Agirre et al. Acta. Cryst. D79, 449-461 (2023) “The CCP4 suite: integrative software for macromolecular crystallography” [doi:10.1107/S2059798323003595]
- Specific CCP4 program reference info: https://www.ccp4.ac.uk/ccp4-v7-0-program-references/
PyMOL
- PyMOL page: https://pymol.org/
- PyMOL wiki: https://pymolwiki.org/index.php/Main_Page
- PyMOL citation info (bottom of page): https://www.pymol.org/support.html?
CCTBX
- CCTBX socumentation: https://cci.lbl.gov/docs/cctbx/
- CCTBX github: https://github.com/cctbx/cctbx_project
- CCTBX reference: Grosse-Kunstleve, R.W., Sauter, N.K., Moriarty, N.W. and Adams, P.D., 2002. The Computational Crystallography Toolbox: crystallographic algorithms in a reusable software framework. Applied Crystallography, 35(1), pp.126-136. https://doi.org/10.1107/S0021889801017824